Section: New Results
CNuclei/cytoplasm detection and classification in genome-wide RNAi screens
Participants : Eric Debreuve, Djampa Kozlowski, Florence Besse, Xavier Descombes.
This work is made in collaboration with Fabienne De Graeve (iBV).
The work described hereafter is part of the RNAGRIMP ANR project which started in January 2016 and lasts 48 months (see Section 7.2.1). A pilot genome-wide RNAi (Ribonucleic Acid interference) screen on Drosophila cultured cells has been performed with different mutant conditions. The purpose is to study the density and repartition of cytoplasmic RNP (RiboNucleoprotein Particles) granules containing the IMP protein (IGF-II mRNA-binding protein where IGF stands for Insulin-like Growth Factor).
Two series of images have been acquired using fluorescence microscopy: one where the cell cytoplasm has been stained with GFP (Green Fluorescent Protein), the second where the nuclei have been stained with DAPI (4',6-diamidino-2-phenylindole). A first task that must be accomplished is to detect the nuclei on the DAPI images, and to learn a classification procedure into living cell or dead cell based on morphologic and radiometric nuclei properties. A CellProfiler (http://cellprofiler.org) pipeline has been developed to automatically detect the nuclei and compute some properties on them. The detection was based on the following main steps: intensity re-scaling, Kapur-based thresholding, and small object discarding. For each detected nucleus, the computed properties were (non-exhaustive list) average intensity, area, granularity, circularity ...
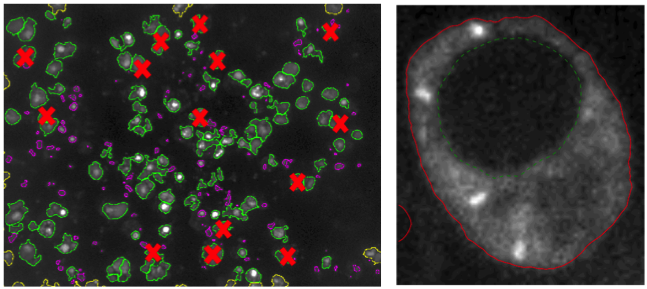
Then, a learning set has been built where a significant number of nuclei were manually assigned their correct (living or dead) class by a biologist of the team. This learning set was fed to CellProfiler Analyst (http://cellprofiler.org/cp-analyst) in order to learn a decision tree for automatic nuclei (hence, cell) classification (see Fig 7, left).
|
Once the living cell nuclei have been identified on the DAPI images, the next step is to segment (i.e., extract automatically the region of) their cytoplasm on the GFP images. Indeed, the target RNP-IMP granules appear in that compartment of the cell and are visible through their GFP response. We are developing an active contour-based segmentation method relying on local image contrast. The current version still has to be robustified in order to be applicable batchwise (see Fig 7, right).